The study of the human microbiome is having an increasing presence and impact on biomedical research projects. Although one of the best described microbiota to date is the intestinal microbiome , which has captured the attention of the scientific community due, among other things, to its high cell density, we must not lose sight of the interest that the study of other microbiota of clinical interest such as nasal, oral, urogenital and / or cutaneous.
In this post we will focus on analyzing the options to stabilize samples of the oral microbiome , which represents approximately 26% of the total human microbiome, and which is attracting the interest of a growing number of researchers in the field of cell biology, microbiology and immunology due to the increasingly evident contribution of this microbiome to states of health and disease not only at the level of the oral cavity (gingivitis, periodontitis …), but also at the systemic level in brain, liver and lung diseases.
The Importance Of Stabilizing Oral Microbiome Samples
Once the microbiome sample is collected, it is essential to proceed to its immediate stabilization to avoid overgrowth and / or degradation of certain populations, and thus guarantee that the result of the subsequent analysis is a faithful reflection of the state and real composition in vivo at the time of collection, ensuring that the obtained microbiological profile represents the phenotype of interest.
Given that the cell cycle of many microorganisms can be around 40 minutes, unless there is a reliable method to stabilize samples of the oral microbiome at the time of collection, the reproducibility and comparability of the microbial profiles between different collection sites or between different studies, it would be practically impossible.
Traditionally, these samples are usually refrigerated or even frozen in order to avoid bacterial growth and degradation, but the truth is that this practice hinders the logistics of the studies and makes the processes more expensive, having to pay maximum attention to maintaining the cold chain. from the collection, transportation, storage and processing of samples.
In order to avoid the need to be subject to the maintenance of this cold chain, and to optimize the quality of the samples, the Canadian DNA Genotek has developed a new battery of kits that allow the collection and stabilization of oral microbiome samples at room temperature during extended periods of time.
Features And Benefits Of Kits To Stabilize Oral Microbiome Samples
- By obtaining reliable profiles of the microbiota at the time of collection, they minimize noise in your data analysis.
- They improve patient / donor adherence through an intuitive and friendly collection method.
- Immediate stabilization of the sample minimizes microbial growth and DNA degradation.
- Eliminates the costs associated with the cold chain in shipping and warehousing.
- Optimize sample handling.
- Maintains the integrity of nucleic acids despite fluctuations in ambient temperature.
Below we summarize the different formats of the kits to stabilize oral microbiome samples and their respective characteristics:
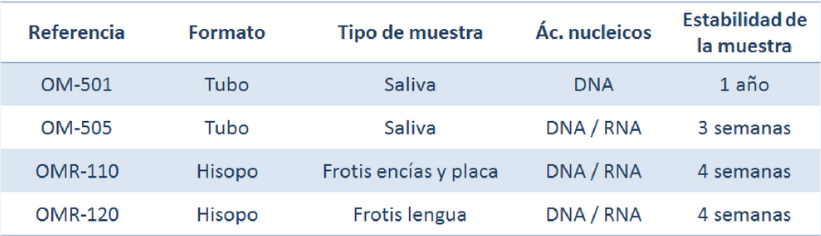
All of these kits eliminate the need for cold chain shipping and warehousing, are tailored for high-throughput processes, and are suitable for NGS applications.
In short, the study of the oral microbiome requires the correct collection and stabilization of the samples to avoid bacterial overgrowth and DNA degradation, guaranteeing the subsequent obtaining of reliable results. Maintaining a cold chain during sample transport and storage can result in additional costs and difficulties in handling the samples, making kits that allow sample collection and stabilization at room temperature a great alternative. booming among researchers.